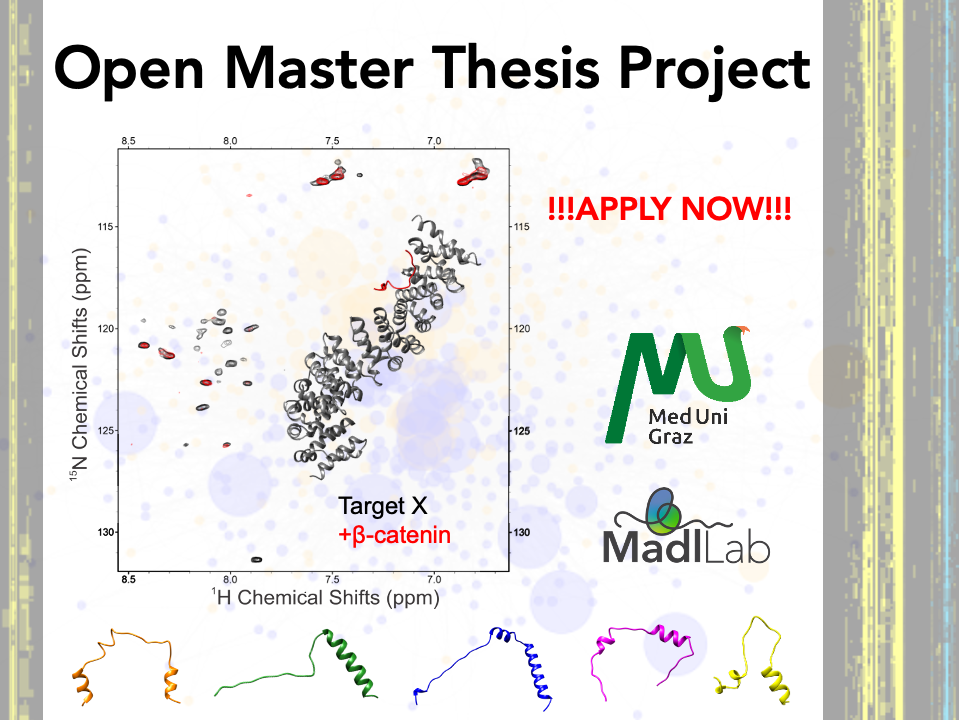
The Madl lab conducts research in the field of structural biology and metabolomics with a special focus on signal transduction and (mis)regulation in human diseases and ageing. More specifically, we investigate the molecular mechanisms how disordered proteins mediate signal transfer via an intricate network of protein interactions and post-translational modifications. We work at the interface of structural biology, biophysics, cell biology, and medicine to study the molecular mechanisms linking key regulatory processes involved in signaling and metabolism. Nuclear magnetic resonance (NMR) spectroscopy and biophysical methods are the primary tools employed in our laboratory to study the structural and dynamical properties of biomolecules and their interactions under native-like solution conditions.
We are located at Otto Loewi Research Center, Medical University of Graz, Austria
Here you can find links to the organizations, initiatives and PhD programs we are part of.